Following a very old post (link), and questions from Matthias and Kevin, I’ve finally managed to test the R2-related scripts I wrote long-long time ago…
I’m really sorry, but don’t quite have the time now to really document all functions/actions, but it should be quite straightforward. This script assumes you start from a ABEM S4K file, or with a ABEM AMP file converted using the command ‘s 4kconv.exe -F:a -z0 -x myinputfile.s4k’. If you have the S4K file, you should be able to convert is on the fly with the script (look for the “convert=False” flag). There are no fancy python imports, except the classic trio: numpy, scipy and matplotlib.
NOTE: there seems to be a problem with the os.spawnl() call to R2.exe within the code, I don’t quite know why for now. So, to run the test, I simply launched R2.exe in console and re-ran the python script to plot the results.
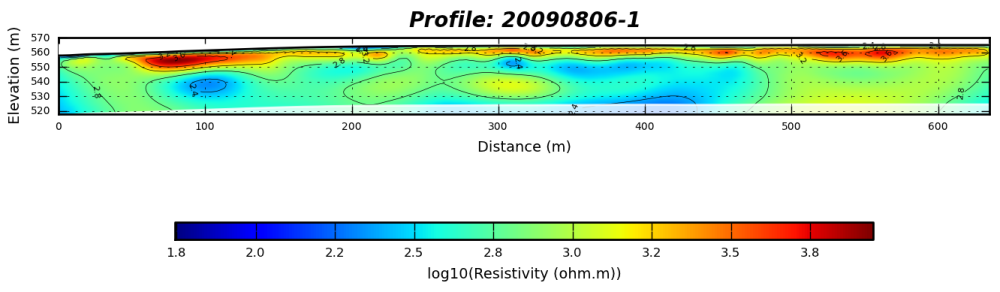
The ouptut of the test case should look like :
The scripts, thus, expects :
- a data folder with
- a .S4K of .AMP file
- a .wgs84 file, N lines of ‘LON LAT ALT’, 1 per electrode
- R2.exe
- if you need to convert, a folder with the s4kconv.exe program
This code should really be improved to be more “dynamic”, if you edit/modify/improve it, please report back and I’ll update/add your contribution to this blog.
The full code and the example data files are present after the break !
The following files should be placed under a /data folder (and remember to copy the R2.exe to this folder too): data.rar
The full code is:
#
import os
import datetime
import sys
from numpy import *
from matplotlib.pyplot import plot, show, scatter
class ResistivityData:
def __init__(self):
self.title = ''
def ParseAMP(self, file):
file = file[:-3] + 'AMP'
f = open(file,'r')
datalines = f.readlines()
f.close()
self.basefolder = os.path.split(file)[0]
# read header
file = os.path.basename(datalines[0].split(":",1)[1].strip())
instrument_model, instrument_serial = datalines[1].split(":",1)[1].strip().split()
date, time = datalines[2].split(":",1)[1].strip().split()
day, month, year = [int(str) for str in date.split("/")]
hours, mins, secs = [int(str) for str in time.split(":")]
# date and time is sometimes completely off
try:
date_time = datetime.datetime(year, month, day, hours, mins, secs)
except ValueError:
print "Warning: date and/or time out of range!"
S4Kfile = AMPfile[:-4] + '.S4K'
if os.path.exists(S4Kfile):
print "Using ctime of file %s" % S4Kfile
ctime = os.path.getctime(S4Kfile)
else:
print "Using ctime of file %s" % AMPfile
ctime = os.path.getctime(AMPfile)
date_time = datetime.datetime.fromtimestamp(ctime)
basestation_coords = datalines[3].split(":",1)[1].strip().split()
header_rows, data_rows, topo_rows = [int(w) for w in datalines[4].split(":",1)[1].strip().split()]
acquisition_mode = datalines[5].split(":",1)[1].strip()
method = datalines[6].split(":",1)[1].strip()
electrode_layout = int(datalines[7].split(":",1)[1].strip().split()[0])
coordinate_type = datalines[8].split(":",1)[1].strip()
dE = float(datalines[9].split(":",1)[1].strip())
self.dE = dE
PPlus = []
PMinus = []
CPlus = []
CMinus = []
Resist = []
for i, line in enumerate(datalines[27:]):
no, time, ax, ay, az, bx, by, bz, mx, my, mz, nx, ny, nz, i, v, r, error = line.split()
# print no, int(ax)+32, int(bx)+32, int(mx)+32, int(nx)+32, i, v, r, error
p_plus = int(mx)+32
p_minus = int(nx)+32
c_plus = int(ax)+32
c_minus = int(bx)+32
r = float(r)
PPlus.append(p_plus)
PMinus.append(p_minus)
CPlus.append(c_plus)
CMinus.append(c_minus)
Resist.append(r)
# print no, p_plus, p_minus, c_plus, c_minus, r
self.PPlus = PPlus
self.PMinus = PMinus
self.CPlus = CPlus
self.CMinus = CMinus
self.Resist = Resist
def create_mesh(self):
electrode_spacing = self.dE
max_depth = 40.0
layers_thickness = 1.0
mesh_X_increment = electrode_spacing / self.split_factor
### MESH GENERATION :
borders = array([1,1,1,1,1,1,1,2,4,5,10,25,50])
# print sum(borders)
before = borders[::-1]*mesh_X_increment
# print before
after = borders*mesh_X_increment
# print after
central = ones((self.num_electrodes*self.split_factor)+1-self.split_factor-1)*mesh_X_increment
# print 'len central', len(central)
# print 'central', central
tempX = hstack((before,central,after[:-1]))
# print 'len tempX', len(tempX)
# print 'tempX', tempX
old_value = -103.0 * mesh_X_increment # TO CHANGE !!
meshX = []
for value in tempX:
old_value += value
meshX.append(old_value)
# print len(meshX), meshX
bottom = array([2,2,2,2,2,2,2,2,4,8,10,20,50,100])
depth = ones(max_depth / layers_thickness)
tempY = hstack((depth,bottom))
old_value = 0.0
meshY = [old_value,]
for value in tempY:
old_value += value
meshY.append(old_value)
topog = zeros(len(meshX))
num_of_elements = (len(meshX)-1)*(len(meshY)-1)
self.meshX = meshX
self.meshY = meshY
self.topog = topog
self.num_of_elements = num_of_elements
def export_R2in(self):
output = os.path.join(self.basefolder,'R2.in')
output = open(output,'w')
num_electrodes = self.num_electrodes
split_factor = self.split_factor
output.write("Header generated\n")
output.write("\n")
output.write(" 1 4 3.0 0 1\n")
output.write("\n")
output.write(" %i %i << numnp_x, numnp_y\n" % (len(self.meshX),len(self.meshY)))
output.write("\n")
out = ""
for i, value in enumerate(self.meshX):
if i % 15 == 0:
output.write("%s\n"% out)
out = ""
out += "%f\t"%value
output.write("%s\n"% out)
output.write("\n")
out = ""
for i, value in enumerate(self.topog):
if i % 15 == 0:
output.write("%s\n"% out)
out = ""
out += "%f\t"%value
output.write("%s\n"% out)
output.write("\n")
out = ""
for i, value in enumerate(self.meshY):
if i % 15 == 0:
output.write("%s\n"% out)
out = ""
out += "%f\t"%value
output.write("%s\n"% out)
output.write("""
1 << num_regions
1 %i 100.000 << elem_1,elem_2,value
1 1 << no. patches in x, no. patches in z
1 << inverse_type
1 0 << data type (0=normal;1=log), regularization type
1.0 10 2 1.0 << tolerance, max_iterations, error_mod, alpha_aniso
0.00100 0.02000 -10000.0 10000.0 << a_wgt, b_wgt, rho_min, rho_max
""" % (self.num_of_elements))
electrode_indexes = linspace(0,num_electrodes*split_factor,num_electrodes+1)
electrode_indexes += 13 + split_factor
output.write(" %i << num_electrodes\n" % (num_electrodes))
for i, value in enumerate(electrode_indexes[:-1]):
output.write("%i\t%i\t%i\n" % (i+1, value, 1))
def export_R2Protocoldat(self):
output = os.path.join(self.basefolder,"protocol.dat")
output = open(output,'w')
output.write("%i\n" % len(self.Resist))
i = 1
for P,p,C,c,r in zip(self.PPlus,self.PMinus,self.CPlus,self.CMinus,self.Resist):
output.write("%i\t%i\t%i\t%i\t%i\t%f\n" % (i, P,p,C,c,r))
i+=1
def add_topo(self,file=None):
if file:
f = open(file,'r')
lines = f.readlines()
f.close()
Z = []
for line in lines:
# ID, x, y, z = line.split()
x, y, z = line.split()
z = float(z)
Z.append(z)
x_old = linspace(1.,self.num_electrodes, self.num_electrodes)
x_new = linspace(1.,self.num_electrodes, (self.num_electrodes*self.split_factor)+1-self.split_factor-1)
if file:
Z = array(Z)
Z_new = interp(x_new,x_old,Z)
else:
Z_new = ones(len(x_new))*100.
before = linspace(Z_new[0],Z_new[0],13)
after = linspace(Z_new[-1],Z_new[-1],12)
Z_new = hstack((before,Z_new, after))
before = linspace(-12,0,13)
after = linspace(self.num_electrodes+1,self.num_electrodes+13, 13)
x_new = hstack((before, x_new, after))
self.topog = Z_new
class R2_Data:
def __init__(self, num_electrodes, split_factor, dE):
self.num_electrodes = num_electrodes
self.split_factor = split_factor
self.dE = dE
self.title = ''
def extract_topo(self, xin, yin, Rin,Show=False):
#EXTRACT THE TOPOGRAPHY, hence THE Max yin for each xin
last_xi = 9999
Yi = []
Xi = []
for xi,yi in zip(xin,yin):
if xi != last_xi:
Yi.append(yi)
Xi.append(xi)
last_xi = xi
topoy = array(Yi)
topox = array(Xi)
if Show:
plt.scatter(xin,yin,marker='+')
plt.scatter(topox,topoy)
plt.grid(True)
show()
return topox, topoy
def read_result(self,filename=None,title='UnNamedProfile',maxdepth=None):
if filename:
input = filename
else:
input = os.path.join(self.basefolder,"f001_res.dat")
if not maxdepth:
maxdepth = 40
input = open(input,'r')
lines = input.readlines()
input.close()
xin = []
yin = []
Rin = []
for line in lines:
x,y,r,logr = line[:-1].split()
xin.append(float(x))
yin.append(float(y))
Rin.append(float(logr))
from matplotlib.mlab import griddata
from numpy import linspace
from scipy import arange, array, meshgrid, ma
from matplotlib import cm
import matplotlib.pyplot as plt
xin = array(xin)
yin = array(yin)
Rin = array(Rin)
topox, topoy = self.extract_topo(xin, yin, Rin)
xi = linspace(0.,(self.num_electrodes-1)*self.dE,250)
# testing
ymax = max(topoy)
ymin = min(topoy) - maxdepth
yi = linspace(ymin,ymax,maxdepth+1)
xi2, yi2 = meshgrid(xi,yi)
print 'yin', yin
zi = griddata(xin,yin,Rin,xi2,yi2)
print 'zi',zi
extent = (min(xi),max(xi),min(yi),max(yi))
plt.figure()
ax = plt.subplot(111,aspect=2.5)
sc = plt.imshow(zi, cmap=cm.jet,interpolation='bicubic',origin='lower',extent=extent,zorder=-10)
cs = plt.contour(zi,origin='lower',extent=extent,colors='k',linewidths=0.25,antialiased=True,zorder=-5)
plt.clabel(cs, fontsize=4, inline=1,fmt='%.1f')
# print ax.get_units()
x1 = topox[11:-11]
y1 = topoy[11:-11]
y2 = ones(len(topoy[11:-11]))*max(topoy)
plt.fill_between(x1,y1+.05,y2,color='w',zorder=10)
plt.fill_between(x1,y1-maxdepth-.5,color='w',zorder=-1,alpha=0.8)
cb = plt.colorbar(sc, orientation="horizontal", spacing="uniform", format="%.1f",aspect=40,shrink=0.75)
cb.set_label('log10(Resistivity (ohm.m))',fontsize=7)
# plt.plot(linspace(0,max(xi),len(self.topog[13:-13])),self.topog[13:-13],'k-')
plt.plot(x1,y1,'k-',zorder=60)
for t in cb.ax.get_xticklabels():
t.set_fontsize(6)
if self.title == '':
self.title = "Profile: " + title
plt.title(self.title,fontsize=10, style='italic', weight='bold')
plt.xticks(fontsize=5)
plt.yticks(fontsize=5)
plt.ylabel('Elevation (m)',fontsize=7)
plt.xlabel('Distance (m)',fontsize=7)
filename = os.path.join(self.basefolder,title + '.R2.PNG')
plt.grid(color='k',linestyle=':',linewidth=0.3,alpha=0.9,zorder=2)
plt.xlim(0,(self.num_electrodes-1)*self.dE)
plt.ylim(min(yi),max(yi)+5)
# plt.savefig(filename,dpi=200,orientation='landscape',papertype='a4')
plt.savefig(filename,dpi=200)
plt.show()
# plt.clf()
def s4kconv(s4kfile):
"""
Wrapper for SAS4000 S4KCONV program (DOS!)
"""
"""
# convert path to DOS path using cygwin
cmd = r'cygpath -d "%s"' % s4kfile.replace('\\', '/')
stdin, stdout = os.popen2(cmd, 't')
stdin.write('\n')
stdin.close()
dos_s4kfile = stdout.readline()
dos_s4kfile = dos_s4kfile.strip()
stdout.close()
# convert program path to DOS as well
cmd = r'cygpath -d "%s"' % os.path.join(SAS4000_DIR, "s4kconv.exe").replace('\\', '/')
stdin, stdout = os.popen2(cmd, 't')
stdin.write('\n')
stdin.close()
dos_s4kconv_path = stdout.readline()
dos_s4kconv_path = dos_s4kconv_path.strip()
stdout.close()
"""
## SAS4000 path
SAS4000_DIR = os.path.abspath("./SAS4000")
PROTOCOL_DIR = os.path.join(SAS4000_DIR, "C2P2")
dos_s4kfile = s4kfile
dos_ampfile = dos_s4kfile[:-4] + ".AMP"
if os.path.exists(dos_ampfile):
os.unlink(dos_ampfile)
dos_s4kconv_path = os.path.join(SAS4000_DIR, "s4kconv.exe")
#args = r'%s %s %s' % (dos_s4kconv_path, '-F:a', dos_s4kfile)
#os.spawnle(os.P_WAIT, os.path.join(SAS4000_DIR, "s4kconv"), args, os.environ)
os.spawnl(os.P_WAIT, os.path.join(SAS4000_DIR, "s4kconv.exe"), dos_s4kconv_path, '-F:a -z0 -x', dos_s4kfile)
tmpfile = dos_ampfile[:-4] + ".TMP"
ampfile = s4kfile[:-4] + ".AMP"
os.rename(dos_ampfile, tmpfile)
fin = open(tmpfile, 'r')
fout = open(ampfile, 'w')
lines = fin.readlines()
fin.close()
fout.write("Filename: %s\n" % os.path.split(s4kfile)[1])
for line in lines[1:]:
fout.write(line)
fout.close()
os.unlink(tmpfile)
def test_run():
# file = r"c:\Vecquee01.s4k"
file = os.path.abspath(r"data\20090806-1.AMP")
# title = 'Crossing Railway - 2009-08-20'
title = '20090806-1'
has_topo = True
work_on_AMP = True
convert = False
run = True
num_electrodes = 128
split_factor = 1.0
dE = 5
if work_on_AMP and convert:
s4kconv(file)
data = ResistivityData()
data.num_electrodes = num_electrodes
data.split_factor = split_factor
data.dE = dE
if work_on_AMP:
data.ParseAMP(file)
else:
# you have to define the arrays by providing the values :
# data.CMinus = array()
# data.CPlus = array()
# data.PMinus = array()
# data.PPlus = array()
# data.Resist = array()
pass
data.create_mesh() #is based on the topo file
if has_topo:
topo_file = file[:-3] + 'wgs84'
data.add_topo(topo_file)
data.export_R2Protocoldat()
data.export_R2in()
print "AT THIS POINT ; LAUNCH R2.EXE IN THE DATA FOLDER, THEN RERUN THIS SCRIPT TO PLOT THE RESULT"
# if run:
# os.chdir(os.path.split(file)[0])
# print os.path.join(os.path.split(file)[0],'R2.exe')
# os.spawnl(os.P_WAIT, os.path.join(os.path.split(file)[0],'R2.exe'))
Result = R2_Data(num_electrodes=num_electrodes,split_factor=split_factor,dE=dE)
if work_on_AMP:
Result.basefolder = data.basefolder
Result.read_result(title=title)
else:
#Result.read_result(filename="PATH_TO_ANOTHER_FILE",title=title)
pass
if __name__ == "__main__":
test_run()

Hey Thanks!
this gives a great starting point for our workflow using R2
My pleasure ! (btw I’m interested in the workflow !)
Hello!
I am wondering to write such program I’m Matlab, but I dont understand what kind of data I should use for that, having dat file from field data only.
Can you help me?
Hi,
No, I can’t help. First, I use only matlab when I need to translate code to Python, this program is fairly simple, but I’d suggest you go ahead and learn python, it’s much more fun ! Good luck !
One More question, please!
Do you use inversion in this program!? Is it least square inversion?
Griddata interpolation was used here, but how can you evaluate the inversion results?